رؤى - Computational Biology - # Microanatomical and Genetic Profiling of Pancreatic Intraepithelial Neoplasias (PanINs)
Comprehensive 3D Mapping Reveals Extensive Multifocality and Genetic Heterogeneity of Pancreatic Precancerous Lesions in Humans
المفاهيم الأساسية
Pancreatic intraepithelial neoplasias (PanINs), the most common precursors of pancreatic cancer, exhibit extensive multifocality and genetic heterogeneity within the normal human pancreas.
الملخص
The study provides a comprehensive microanatomical survey and 3D genomic mapping of human pancreatic intraepithelial neoplasias (PanINs), the most common precursors of pancreatic cancer. Key insights:
- The researchers analyzed 46 large samples of grossly normal human pancreas using a machine-learning pipeline for quantitative 3D histological reconstruction at single-cell resolution.
- They found a mean burden of 13 PanINs per cm3 of pancreatic tissue and extrapolated that the normal intact adult pancreas harbors hundreds of PanINs, almost all with oncogenic KRAS hotspot mutations.
- Most PanINs originate as independent clones with distinct somatic mutation profiles, indicating multifocal origins.
- Some spatially continuous PanINs were found to contain multiple KRAS mutations, with computational and in situ analyses demonstrating that different KRAS mutations localize to distinct cell subpopulations within these neoplasms, indicating polyclonal origins.
- The extensive multifocality and genetic heterogeneity of PanINs raises important questions about the mechanisms driving precancer initiation and conferring differential progression risk in the human pancreas.
- This detailed 3D genomic mapping of molecular alterations in human PanINs provides an empirical foundation for early detection and rational interception of pancreatic cancer.
تخصيص الملخص
إعادة الكتابة بالذكاء الاصطناعي
إنشاء الاستشهادات
ترجمة المصدر
إلى لغة أخرى
إنشاء خريطة ذهنية
من محتوى المصدر
زيارة المصدر
www.nature.com
3D genomic mapping reveals multifocality of human pancreatic precancers - Nature
الإحصائيات
The normal intact adult pancreas harbors hundreds of PanINs, almost all with oncogenic KRAS hotspot mutations.
The researchers found a mean burden of 13 PanINs per cm3 of pancreatic tissue.
اقتباسات
"We found that most PanINs originate as independent clones with distinct somatic mutation profiles."
"Some spatially continuous PanINs were found to contain multiple KRAS mutations; computational and in situ analyses demonstrated that different KRAS mutations localize to distinct cell subpopulations within these neoplasms, indicating their polyclonal origins."
الرؤى الأساسية المستخلصة من
by Alicia M. Br... في www.nature.com 05-01-2024
https://www.nature.com/articles/s41586-024-07359-3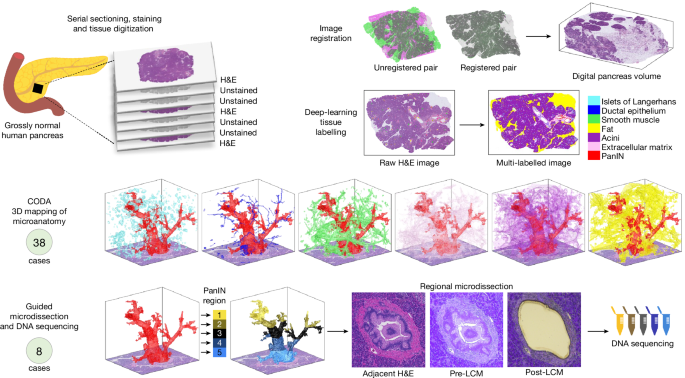
استفسارات أعمق
What are the potential mechanisms driving the extensive multifocality and genetic heterogeneity of PanINs within the normal human pancreas?
The extensive multifocality and genetic heterogeneity of PanINs in the normal human pancreas can be attributed to several potential mechanisms. Firstly, the presence of oncogenic KRAS hotspot mutations in almost all PanINs indicates a common driver mutation that initiates these precancerous lesions. This suggests that the clonal expansion of cells harboring KRAS mutations contributes to the multifocality of PanINs. Additionally, the independent clonal origins of most PanINs with distinct somatic mutation profiles suggest that these lesions arise from different precursor cells, leading to genetic heterogeneity within the PanIN population. Furthermore, the identification of spatially continuous PanINs containing multiple KRAS mutations indicates that these neoplasms may arise from polyclonal origins, where different cell subpopulations within the lesion harbor distinct mutations. This genetic diversity and clonal evolution contribute to the multifocality and genetic heterogeneity observed in PanINs within the normal human pancreas.
How can the insights from this 3D genomic mapping of PanINs be leveraged to improve early detection and rational interception strategies for pancreatic cancer?
The insights gained from the 3D genomic mapping of PanINs offer valuable opportunities to enhance early detection and interception strategies for pancreatic cancer. By quantifying the burden of PanINs in the normal pancreas and identifying the prevalence of oncogenic KRAS mutations in these lesions, clinicians can develop screening protocols to detect and monitor individuals at high risk for pancreatic cancer. The detailed molecular characterization of PanINs can aid in the identification of specific genetic alterations associated with disease progression, enabling the development of targeted therapies for early intervention. Furthermore, understanding the spatial distribution and clonal origins of PanINs can guide the design of precision medicine approaches that target distinct cell populations within these lesions. Leveraging this 3D genomic mapping data can lead to the development of personalized screening strategies and novel therapeutic interventions aimed at intercepting pancreatic cancer at its earliest stages.
What are the implications of the polyclonal origins of some PanINs for our understanding of the evolutionary dynamics of pancreatic cancer development?
The polyclonal origins of some PanINs have significant implications for our understanding of the evolutionary dynamics of pancreatic cancer development. The presence of multiple KRAS mutations within spatially continuous PanINs suggests that these lesions may arise from the clonal expansion of distinct cell populations with different genetic alterations. This polyclonality indicates that PanINs can evolve through the accumulation of diverse mutations in independent precursor cells, leading to intratumoral heterogeneity. The coexistence of multiple subclones within a single PanIN lesion highlights the complex evolutionary trajectories of pancreatic cancer development, where different cell populations with unique genetic profiles contribute to tumor progression. Understanding the polyclonal origins of PanINs provides insights into the clonal evolution and diversification of pancreatic cancer, emphasizing the need for comprehensive molecular profiling and targeted therapies that address the genetic heterogeneity within these precancerous lesions.
0
