Harnessing Protein Transport Coupling to Achieve Targeted Protein Relocalization for Therapeutic Potential
The article discusses the concept of targeted protein relocalization as a potential therapeutic approach for diseases such as cancer and neurodegenerative disorders. Subcellular protein localization is crucial for regulating protein function, and its disruption can contribute to disease pathogenesis.
The authors identify a collection of shuttle proteins with potent ligands that can be incorporated into targeted relocalization-activating molecules (TRAMs). These TRAMs can be used to relocalize endogenous proteins by coupling them to the trafficking of the shuttle proteins. The authors demonstrate the feasibility of this approach by modulating the steady-state localization of various proteins, including disease-driving mutant proteins such as SMARCB1Q318X, TDP43ΔNLS, and FUSR495X.
The article highlights the use of nuclear hormone receptors as shuttles to redistribute the disease-driving mutant proteins. The TRAM-mediated relocalization of FUSR495X to the nucleus from the cytoplasm was found to correlate with a reduction in the number of stress granules in a model of cellular stress.
The authors also demonstrate the relocalization of endogenous proteins, such as PRMT9, SOS1, and FKBP12, using methionyl aminopeptidase 2 and poly(ADP-ribose) polymerase 1 as endogenous cytoplasmic and nuclear shuttles, respectively.
Furthermore, the article discusses the potential of small-molecule-mediated redistribution of nicotinamide nucleotide adenylyltransferase 1 from nuclei to axons in primary neurons, which was able to slow axonal degeneration and pharmacologically mimic the genetic WldS gain-of-function phenotype in mice resistant to certain types of neurodegeneration.
The concept of targeted protein relocalization could inspire new approaches for treating diseases through interactome rewiring, offering a promising avenue for therapeutic development.
התאם אישית סיכום
כתוב מחדש עם AI
צור ציטוטים
תרגם מקור
לשפה אחרת
צור מפת חשיבה
מתוכן המקור
עבור למקור
www.nature.com
Targeted protein relocalization via protein transport coupling - Nature
תובנות מפתח מזוקקות מ:
by Christine S.... ב- www.nature.com 09-18-2024
https://www.nature.com/articles/s41586-024-07950-8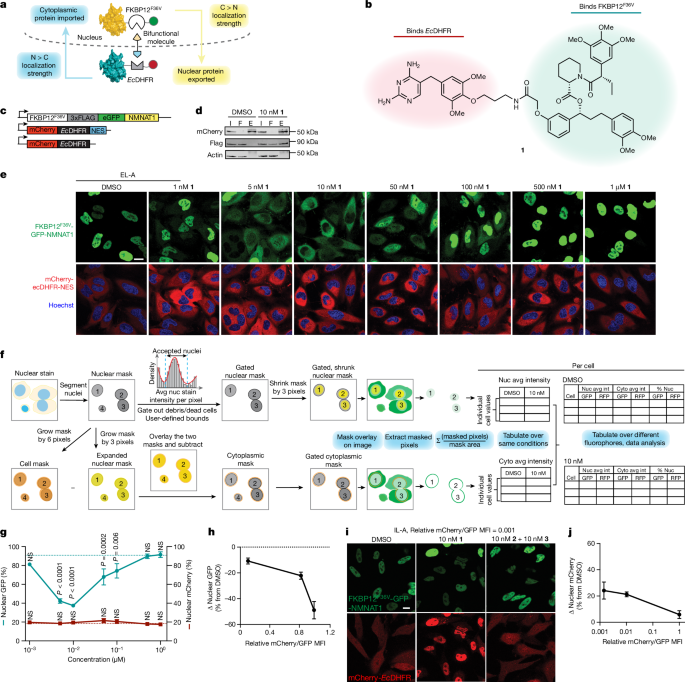
שאלות מעמיקות
