AIRE's Mechanism: Z-DNA and T Cell Tolerization
Keskeiset käsitteet
AIRE relies on Z-DNA and NFE2–MAF to select gene targets for thymic T cell tolerization by enhancing double-stranded break generation and promoter poising.
Tiivistelmä
AIRE, an unconventional transcription factor, influences gene expression in thymic cells to regulate self-reactive T cells. Through Z-DNA and NFE2–MAF, AIRE targets genes with poised promoters, shedding light on mechanisms of T cell tolerance manipulation.
Mukauta tiivistelmää
Kirjoita tekoälyn avulla
Luo viitteet
Käännä lähde
toiselle kielelle
Luo miellekartta
lähdeaineistosta
Siirry lähteeseen
www.nature.com
AIRE relies on Z-DNA to flag gene targets for thymic T cell tolerization - Nature
Tilastot
AIRE enhances the expression of thousands of genes in medullary thymic epithelial cells.
Z-DNA and NFE2–MAF are identified as positive influences on AIRE’s target choices.
Genome-wide mapping studies show that Z-DNA-forming motifs are associated with DNA double-stranded break generation in gene promoters.
Promoters with strong double-stranded break generation are more likely to have poised states with accessible chromatin.
AIRE preferentially targets genes with poised promoters.
Lainaukset
"We propose a model in which Z-DNA anchors the AIRE-mediated transcriptional program by enhancing double-stranded break generation and promoter poising."
Tärkeimmät oivallukset
by Yuan Fang,Ku... klo www.nature.com 03-13-2024
https://www.nature.com/articles/s41586-024-07169-7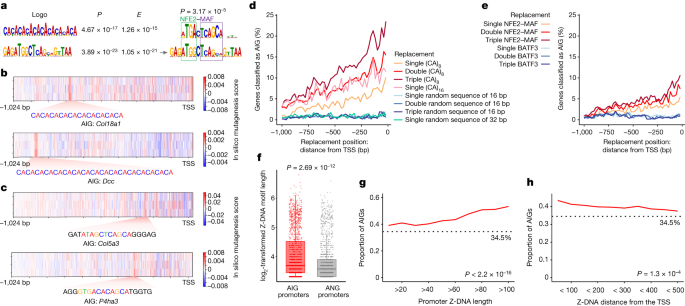
Syvällisempiä Kysymyksiä
How can the discovery of Z-DNA's role in AIRE's mechanism impact future research on autoimmune diseases?
The discovery of Z-DNA's role in AIRE's mechanism could significantly impact future research on autoimmune diseases by providing a new avenue for understanding and potentially manipulating T cell tolerance. By elucidating how AIRE targets specific genes through Z-DNA formation, researchers may uncover novel therapeutic targets for modulating immune responses in autoimmune conditions. This knowledge could lead to the development of more targeted treatments that aim to restore immune tolerance and prevent autoimmunity.
What potential challenges or limitations could arise from manipulating T cell tolerance based on these findings?
Manipulating T cell tolerance based on the findings related to AIRE and Z-DNA may pose several challenges and limitations. One major concern is the potential off-target effects of targeting specific genes involved in immune regulation. Modulating T cell tolerance through interventions aimed at altering gene expression patterns could inadvertently disrupt normal physiological processes, leading to unintended consequences or adverse reactions.
Additionally, the complexity of the immune system and its intricate regulatory mechanisms present a challenge in accurately predicting the outcomes of manipulating T cell tolerance. The interconnected nature of various immune pathways means that interventions targeting one aspect may have cascading effects throughout the immune system, which could be difficult to control or predict.
Ethical considerations regarding genetic manipulation for therapeutic purposes also need to be taken into account when exploring strategies to manipulate T cell tolerance based on these findings. Ensuring safety, efficacy, and long-term implications of such interventions will be crucial before translating these discoveries into clinical applications.
How might understanding the cis-regulatory mechanisms of transcription factors like AIRE contribute to advancements in personalized medicine?
Understanding the cis-regulatory mechanisms of transcription factors like AIRE can pave the way for significant advancements in personalized medicine by enabling tailored approaches for treating autoimmune diseases and other conditions with immunological components. By deciphering how transcription factors regulate gene expression patterns, researchers can identify key molecular targets that drive disease pathogenesis at an individual level.
This personalized approach allows for precision medicine strategies that take into account each patient's unique genetic makeup, environmental influences, and disease manifestations. By leveraging this knowledge, clinicians can develop customized treatment plans that target specific dysregulated pathways implicated by aberrant transcription factor activity such as AIRE-mediated gene expression changes.
Furthermore, insights gained from studying cis-regulatory mechanisms can inform biomarker discovery efforts aimed at stratifying patients based on their molecular profiles or predicting treatment responses more accurately. This enhanced understanding of transcriptional regulation offers opportunities to develop innovative diagnostic tools and therapeutic modalities tailored to individual patients' needs within the realm of personalized medicine.
0
