Proteome-Scale Discovery of Protein Degradation and Stabilization Effectors
Core Concepts
Proteome-scale platform identifies human proteins for targeted protein degradation and stabilization.
Abstract
The content discusses the potential of targeted protein degradation and stabilization as therapeutic modalities, highlighting the limitations in utilizing E3 ligases and deubiquitinases. A synthetic proteome-scale platform was established to identify human proteins that can promote protein degradation or stabilization. The study reveals a diverse range of effectors in the human proteome with varying activities against different targets, showcasing their potential for therapeutic applications.
Key Highlights:
- Targeted protein degradation and stabilization are potent therapeutic strategies.
- Limited utilization of E3 ligases and deubiquitinases hinders approach effectiveness.
- Synthetic proteome-scale platform identifies human proteins for protein modulation.
- Human proteome contains a variety of effectors influencing protein stability.
- Comparison of activities among E3 ligases and deubiquitinases conducted.
- Non-canonical protein degraders and stabilizers characterized.
- Top degraders show higher potency against therapeutically relevant targets than current options.
- Study provides a functional catalogue of stability effectors for targeted protein modulation.
Customize Summary
Rewrite with AI
Generate Citations
Translate Source
To Another Language
Generate MindMap
from source content
Visit Source
www.nature.com
Proteome-scale discovery of protein degradation and stabilization effectors - Nature
Stats
However, only a few of the hundreds of E3 ligases and deubiquitinases in the human proteome have been harnessed for this purpose, which substantially limits the potential of the approach.
Here we established a synthetic proteome-scale platform to functionally identify human proteins that can promote the degradation or stabilization of a target protein in a proximity-dependent manner.
Quotes
"Our study provides a functional catalogue of stability effectors for targeted protein degradation and stabilization."
"The top degraders were more potent against multiple therapeutically relevant targets than the currently used E3 ligases cereblon and VHL."
Key Insights Distilled From
by Juline Poirs... at www.nature.com 03-20-2024
https://www.nature.com/articles/s41586-024-07224-3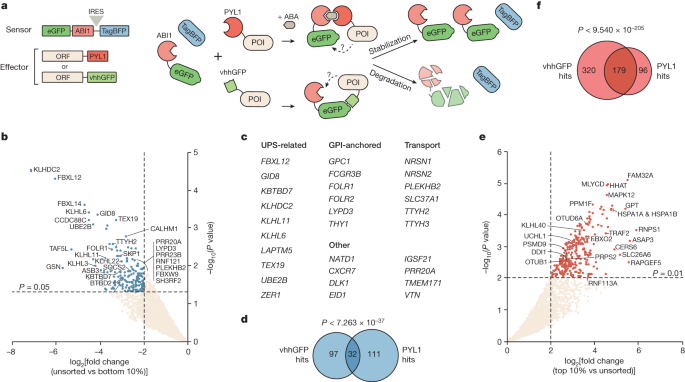
Deeper Inquiries
How can the discovery of new proximity-dependent protein modulators impact drug development
The discovery of new proximity-dependent protein modulators can significantly impact drug development by expanding the druggable target space and offering novel therapeutic modalities. These modulators, which can promote the degradation or stabilization of target proteins in a proximity-dependent manner, provide researchers with a broader toolkit to manipulate protein levels within cells. This approach allows for more precise control over specific targets, potentially leading to enhanced efficacy and reduced off-target effects in drug development.
By identifying and characterizing non-canonical protein degraders and stabilizers through proteome-scale platforms, researchers can uncover previously untapped avenues for therapeutic intervention. This expanded repertoire of effectors opens up opportunities to target proteins that were previously considered undruggable or challenging to modulate pharmacologically. Ultimately, the discovery of new proximity-dependent protein modulators offers a promising strategy to address unmet medical needs and develop innovative treatments for various diseases.
What challenges might arise when utilizing non-canonical protein degraders for therapeutic purposes
When utilizing non-canonical protein degraders for therapeutic purposes, several challenges may arise that need to be carefully addressed. One key challenge is ensuring specificity in targeting the desired proteins while minimizing off-target effects on unintended substrates. Non-canonical effectors may exhibit different substrate preferences or interact with multiple proteins simultaneously, raising concerns about potential cytotoxicity or unintended biological consequences.
Another challenge lies in optimizing the delivery mechanisms of these non-canonical protein degraders to achieve sufficient intracellular concentrations at the desired sites of action. Strategies such as developing appropriate formulations or delivery vehicles that enhance cellular uptake and stability while minimizing systemic exposure will be crucial for effective therapeutic applications.
Additionally, understanding the complex interplay between non-canonical effectors and cellular pathways is essential to predict potential side effects or resistance mechanisms that could emerge during treatment. Comprehensive preclinical studies are necessary to evaluate the safety profile, pharmacokinetics, and pharmacodynamics of these novel therapeutics before advancing them into clinical trials.
How does understanding diverse activities among effectors contribute to personalized medicine approaches
Understanding diverse activities among effectors plays a pivotal role in personalized medicine approaches by enabling tailored treatment strategies based on individual patient characteristics. The identification of human E3 ligases and deubiquitinases with varying activities against diverse targets provides insights into how different patients may respond differently to targeted protein degradation or stabilization therapies.
By comprehensively comparing effector activities across different patient populations, healthcare providers can select optimal treatment options that are most likely to be effective for each individual's unique molecular profile. This personalized approach enhances treatment outcomes by maximizing efficacy while minimizing adverse reactions or ineffectiveness due to genetic variations or other factors influencing effector interactions with target proteins.
Moreover, leveraging this knowledge allows for precision medicine interventions where specific effector profiles can be matched with patient-specific biomarkers or disease signatures to tailor therapy regimens accordingly. By incorporating information on effector diversity into clinical decision-making processes, personalized medicine approaches strive towards more effective treatments that consider each patient's biological nuances and optimize therapeutic outcomes based on individualized responses.
0
