Comprehensive Analysis of Centromeric Variation and Evolution Across Human and Primate Genomes
Core Concepts
Human and primate centromeres exhibit extensive sequence variation, rapid evolution, and dynamic changes in higher-order repeat structures, challenging traditional models of centromere biology.
Abstract
This study provides a comprehensive analysis of human and primate centromeric sequences, revealing previously uncharacterized patterns of variation and evolution. Key findings include:
Comparison of human centromeres from two genomes shows at least a 4.1-fold increase in single-nucleotide variation compared to flanking unique regions, and up to 3-fold variation in size.
45.8% of human centromeric sequences cannot be reliably aligned due to the emergence of new α-satellite higher-order repeats (HORs).
26% of human centromeres differ in their kinetochore position by >500 kb, as determined by DNA methylation and CENP-A chromatin immunoprecipitation.
Sequencing of orthologous centromeres from chimpanzee, orangutan, and macaque genomes reveals a nearly complete turnover of α-satellite HORs, with characteristic species-specific changes.
Phylogenetic analysis supports limited to no recombination between the short (p) and long (q) arms of human centromeres, and suggests a monophyletic origin for novel α-satellite HORs, providing a strategy to estimate the rate of centromeric DNA amplification and mutation.
These findings challenge traditional models of centromere biology and highlight the dynamic and rapidly evolving nature of these essential genomic regions.
The variation and evolution of complete human centromeres - Nature
Stats
At least a 4.1-fold increase in single-nucleotide variation in human centromeres compared to flanking regions
Up to 3-fold variation in size of human centromeres
45.8% of human centromeric sequences cannot be reliably aligned due to new α-satellite higher-order repeats
26% of human centromeres differ in kinetochore position by >500 kb
Quotes
"Human centromeres have been traditionally very difficult to sequence and assemble owing to their repetitive nature and large size1."
"We find that the two sets of centromeres show at least a 4.1-fold increase in single-nucleotide variation when compared with their unique flanks and vary up to 3-fold in size."
"Moreover, we find that 45.8% of centromeric sequence cannot be reliably aligned using standard methods owing to the emergence of new α-satellite higher-order repeats (HORs)."
Key Insights Distilled From
by Glennis A. L... at www.nature.com 04-03-2024
https://www.nature.com/articles/s41586-024-07278-3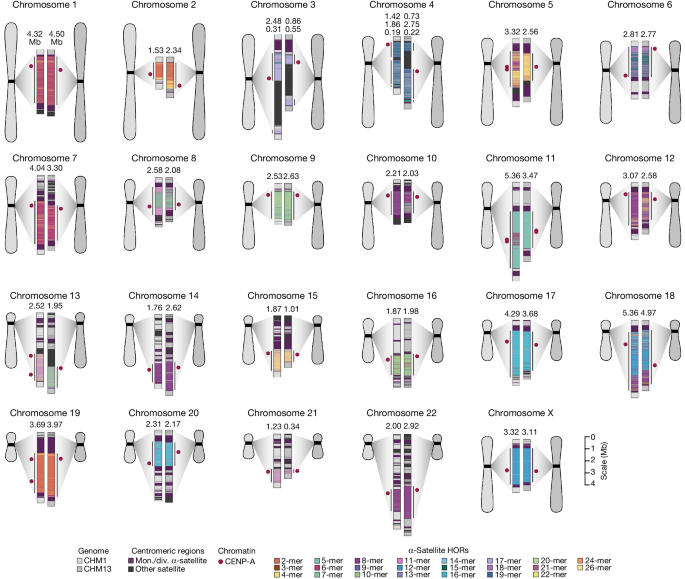
Deeper Inquiries
How do the rapid evolutionary changes in centromeric sequences impact chromosome segregation and genome stability?
The rapid evolutionary changes in centromeric sequences can have significant implications for chromosome segregation and genome stability. Centromeres play a crucial role in ensuring accurate chromosome segregation during cell division by serving as attachment sites for spindle microtubules. Any alterations in centromeric sequences, such as the observed increase in single-nucleotide variation and changes in centromere size, can potentially affect the binding affinity of kinetochore proteins, leading to errors in chromosome segregation. These errors can result in aneuploidy, a condition characterized by an abnormal number of chromosomes in daughter cells, which is a common feature of cancer cells. Additionally, the emergence of new α-satellite higher-order repeats (HORs) that cannot be reliably aligned using standard methods may disrupt the proper assembly of the kinetochore complex, further compromising chromosome segregation fidelity. Overall, the rapid evolutionary changes in centromeric sequences can jeopardize the accurate distribution of genetic material during cell division, impacting genome stability.
What are the potential functional implications of the observed variation in kinetochore positioning within human centromeres?
The observed variation in kinetochore positioning within human centromeres can have several functional implications. Kinetochore positioning is critical for the proper attachment of spindle microtubules during cell division, ensuring accurate chromosome segregation. The variation in kinetochore positioning, as indicated by differences of >500 kb in 26% of centromeres, suggests potential alterations in the spatial organization of centromeric chromatin. This variation may influence the accessibility of kinetochore proteins to centromeric DNA, affecting the stability of kinetochore-microtubule interactions. Changes in kinetochore positioning could also impact the tension exerted on sister chromatids during chromosome alignment, potentially leading to errors in chromosome segregation. Furthermore, alterations in kinetochore positioning may influence the epigenetic regulation of centromeric chromatin, affecting centromere function and stability. Therefore, the observed variation in kinetochore positioning within human centromeres could have profound effects on chromosome segregation accuracy and genome integrity.
Could the dynamic nature of centromeric DNA be exploited for the development of novel genetic tools or therapeutic approaches?
The dynamic nature of centromeric DNA, as evidenced by the turnover of α-satellite higher-order repeats (HORs) and the emergence of novel α-satellite HORs, presents exciting opportunities for the development of novel genetic tools and therapeutic approaches. The unique sequence characteristics and rapid evolution of centromeric DNA could be harnessed for applications such as targeted genome editing and gene therapy. The idiosyncratic changes in α-satellite HORs among different species provide a rich source of genetic diversity that could be leveraged to engineer specific centromere sequences for precise genome modifications. Additionally, the monophyletic origin of novel α-satellite HORs offers insights into the rate of saltatory amplification and mutation of centromeric DNA, which could inform the development of strategies for manipulating centromere structure and function. By exploiting the dynamic nature of centromeric DNA, researchers may uncover new approaches for gene targeting, chromosomal engineering, and therapeutic interventions that could revolutionize the field of genetics and genomics.
0
