インサイト - Computational Biology - # Reprogrammable Recombinase Enzymes for Targeted Genomic Rearrangements
Programmable RNA-Guided Enzymes Enabling Next-Generation Genome Editing
核心概念
Researchers have developed programmable recombinase enzymes guided by RNA molecules to enable precise, large-scale genome editing capabilities.
要約
The content discusses the long-standing goal of biologists to develop methods for rearranging long DNA sequences in genomes. Recombinase and transposase enzymes that can mediate such genomic rearrangements have been studied extensively, but it has been challenging to reprogram these enzymes to precisely target user-specified genomic sites.
The article then highlights two recent papers in Nature that report the characterization of recombinases that are guided by a 'bridge' RNA molecule. This allows the recombinases to be reprogrammed, enabling new genome-editing capabilities. The RNA-guided recombinases can be used to insert, invert, delete or move kilobase-scale DNA sequences to user-specified locations in cells in a single step, overcoming the limitations of previous approaches.
Programmable RNA-guided enzymes for next-generation genome editing
統計
Biologists have long aspired to develop programmable methods to rearrange long DNA sequences in genomes.
Recombinase and transposase enzymes that mediate genomic rearrangements of large sequences have been studied intensively.
It has been challenging to reprogram these enzymes to precisely target user-specified genomic sites.
引用
"A long-standing aspiration for biologists has been to develop programmable methods to rearrange long DNA sequences in genomes."
"Two papers in Nature now report the characterization of recombinases that are guided by a 'bridge' RNA molecule, and can thereby be reprogrammed to enable new genome-editing capabilities."
抽出されたキーインサイト
by Connor J. To... 場所 www.nature.com 06-26-2024
https://www.nature.com/articles/d41586-024-01461-2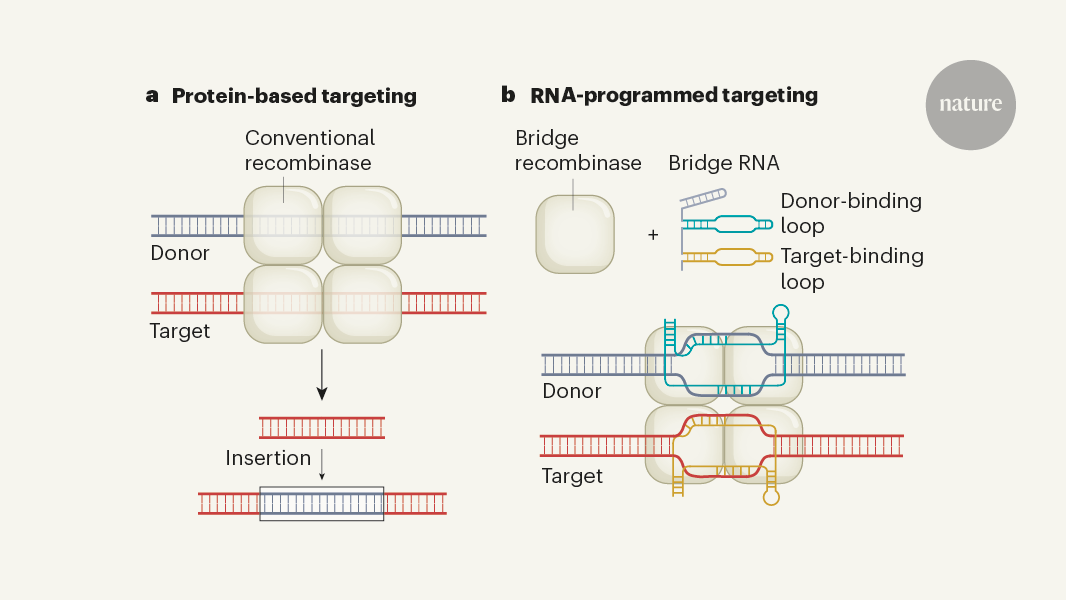
深掘り質問
What are the potential applications of these programmable RNA-guided recombinase enzymes beyond genome editing?
The programmable RNA-guided recombinase enzymes have potential applications beyond genome editing in various fields. One significant application is in synthetic biology, where these enzymes can be utilized to engineer complex genetic circuits and pathways with precise control over DNA rearrangements. This can enable the creation of novel biological systems for biotechnological applications, such as the production of biofuels, pharmaceuticals, and biomaterials. Additionally, these enzymes could be employed in gene therapy to treat genetic disorders by precisely modifying patient genomes to correct mutations or introduce therapeutic genes. Furthermore, the ability to rearrange long DNA sequences in a single step opens up possibilities for studying gene function, regulatory elements, and evolutionary processes in a more targeted and efficient manner.
How do the capabilities of these new enzymes compare to other genome editing tools like CRISPR-Cas9?
The capabilities of programmable RNA-guided recombinase enzymes offer several advantages over other genome editing tools like CRISPR-Cas9. While CRISPR-Cas9 is widely used for precise DNA editing, it primarily functions by inducing double-strand breaks in the DNA, which can lead to unintended mutations and off-target effects. In contrast, the RNA-guided recombinase enzymes can rearrange long DNA sequences without the need for double-strand breaks, potentially reducing the risk of off-target effects and increasing the precision of genome editing. Moreover, these enzymes allow for the insertion, inversion, deletion, or movement of kilobase-scale DNA sequences in a single step, providing a more efficient and versatile approach to genome editing compared to CRISPR-Cas9, which typically requires multiple steps for complex DNA rearrangements.
What technical challenges or limitations still need to be addressed to fully realize the potential of this technology?
Despite the promising capabilities of programmable RNA-guided recombinase enzymes, several technical challenges and limitations need to be addressed to fully realize the potential of this technology. One key challenge is optimizing the efficiency and specificity of the enzyme-mediated DNA rearrangements to ensure accurate targeting of user-specified genomic sites. Improving the delivery methods of these enzymes into target cells and tissues is another hurdle that needs to be overcome to enable widespread applications in gene therapy and biotechnology. Additionally, enhancing the scalability and throughput of the genome editing process using these enzymes will be crucial for their practical implementation in large-scale genetic engineering projects. Furthermore, understanding the potential long-term effects of genome rearrangements mediated by these enzymes on cell viability, stability, and function is essential for ensuring the safety and reliability of this technology in diverse biological contexts. Addressing these technical challenges will be instrumental in unlocking the full potential of programmable RNA-guided recombinase enzymes for next-generation genome editing and beyond.
0
