Enhancing Prime Editing Efficiency through Harnessing an Endogenous RNA-Binding Protein
The content describes research aimed at identifying cellular determinants that influence the performance of prime editing, a genome editing technology that enables precise modifications through reverse transcription of template sequences appended to CRISPR-Cas guide RNAs.
The researchers developed scalable prime editing reporters and performed genome-scale CRISPR-interference screens, which revealed that the small RNA-binding exonuclease protection factor La is a key mediator of prime editing efficiency. Further investigation showed that La promotes prime editing across different approaches (PE2, PE3, PE4, and PE5), edit types (substitutions, insertions, and deletions), endogenous loci, and cell types, but has no consistent effect on genome-editing approaches that rely on standard, unextended guide RNAs.
The researchers found that La functionally interacts with the 3' ends of polyuridylated prime editing guide RNAs (pegRNAs). Leveraging this insight, they developed a prime editor protein (PE7) fused to the RNA-binding, N-terminal domain of La, which improved prime editing performance with expressed pegRNAs, engineered pegRNAs (epegRNAs), and synthetic pegRNAs optimized for La binding.
The findings provide key insights into how prime editing components interact with the cellular environment and suggest general strategies for stabilizing exogenous small RNAs, which can be applied to enhance the efficiency of prime editing and other genome editing technologies.
Customize Summary
Rewrite with AI
Generate Citations
Translate Source
To Another Language
Generate MindMap
from source content
Visit Source
www.nature.com
Improving prime editing with an endogenous small RNA-binding protein - Nature
Kluczowe wnioski z
by Jun Yan,Paul... o www.nature.com 04-03-2024
https://www.nature.com/articles/s41586-024-07259-6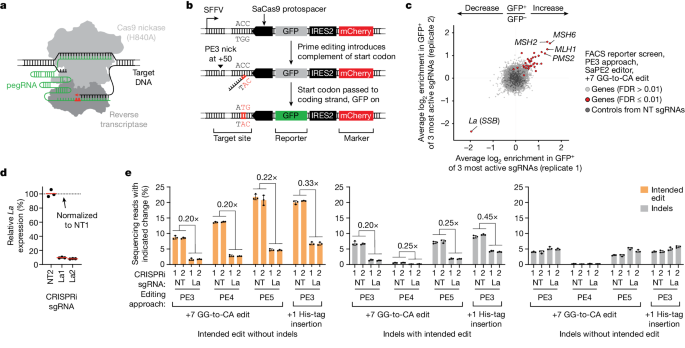
Głębsze pytania
