Structural Insights into the Mechanism of Prime Editor-Mediated Genome Editing
Основные понятия
The prime editor system, composed of Cas9 nickase and engineered reverse transcriptase, utilizes a prime editing guide RNA (pegRNA) to facilitate precise genome editing, but the molecular mechanism was previously unclear. This study provides structural insights into the stepwise process of pegRNA-guided reverse transcription by the prime editor complex.
Аннотация
The prime editor system is a powerful genome editing tool that combines the Streptococcus pyogenes Cas9 nickase (nSpCas9) and an engineered Moloney murine leukemia virus reverse transcriptase (M-MLV RT) to facilitate a wide range of precise genome edits in living cells. However, the molecular mechanism underlying the pegRNA-guided reverse transcription process was not well understood due to a lack of structural information.
This study presents cryo-electron microscopy structures of the SpCas9–M-MLV RTΔRNaseH–pegRNA–target DNA complex in multiple states, including the pre-initiation, initiation, elongation, and termination stages. The key findings are:
The termination structure and functional analysis reveal that M-MLV RT extends reverse transcription beyond the expected site, resulting in the incorporation of scaffold-derived sequences that can cause undesired edits at the target loci.
Structural comparisons among the different states show that M-MLV RT maintains a consistent position relative to SpCas9 during reverse transcription, while the pegRNA-synthesized DNA heteroduplex builds up along the surface of SpCas9.
Based on these structural insights, the researchers rationally engineered pegRNA variants and prime-editor variants in which M-MLV RT is fused within SpCas9, aiming to develop a more versatile prime editing toolbox.
The study provides valuable structural insights into the stepwise mechanism of prime editing, which will contribute to the further development and optimization of this powerful genome editing technology.
Structural basis for pegRNA-guided reverse transcription by a prime editor - Nature
Статистика
The prime editor system is composed of Streptococcus pyogenes Cas9 nickase (nSpCas9) and engineered Moloney murine leukemia virus reverse transcriptase (M-MLV RT).
The cryo-electron microscopy structures were obtained for the SpCas9–M-MLV RTΔRNaseH–pegRNA–target DNA complex in multiple states, including pre-initiation, initiation, elongation, and termination.
Цитаты
"The termination structure, along with our functional analysis, reveals that M-MLV RT extends reverse transcription beyond the expected site, resulting in scaffold-derived incorporations that cause undesired edits at the target loci."
"Structural comparisons among the pre-initiation, initiation and elongation states show that M-MLV RT remains in a consistent position relative to SpCas9 during reverse transcription, whereas the pegRNA–synthesized DNA heteroduplex builds up along the surface of SpCas9."
Ключевые выводы из
by Yutaro Shuto... в www.nature.com 05-29-2024
https://www.nature.com/articles/s41586-024-07497-8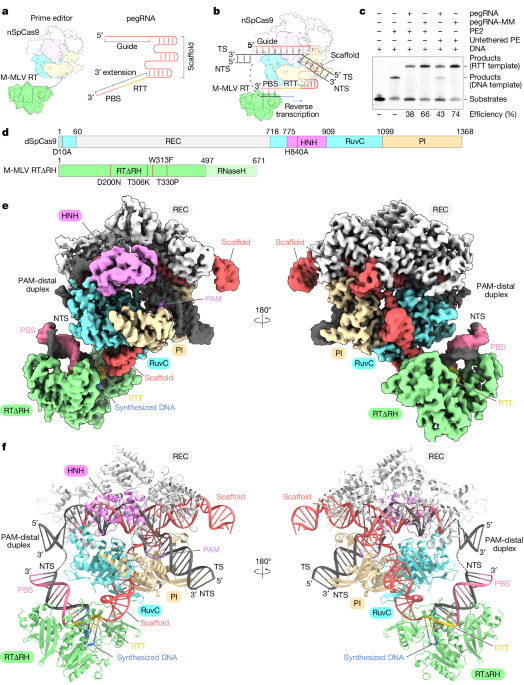
Дополнительные вопросы
How can the undesired scaffold-derived incorporations caused by the extended reverse transcription be further minimized or eliminated to improve the precision of prime editing?
To minimize or eliminate the undesired scaffold-derived incorporations caused by the extended reverse transcription in prime editing, several strategies can be employed. One approach could involve optimizing the design of the pegRNA to enhance its specificity and efficiency. This optimization may include modifying the pegRNA sequence to reduce off-target effects and improve the fidelity of reverse transcription. Additionally, the use of modified nucleotides or chemical modifications in the pegRNA backbone can help stabilize the pegRNA–synthesized DNA heteroduplex and prevent unintended incorporations during reverse transcription. Furthermore, fine-tuning the interaction between the pegRNA and M-MLV RT can also aid in controlling the extension of reverse transcription to the desired site, thereby reducing the occurrence of undesired edits at the target loci. Continuous refinement of the prime editing system through experimental validation and computational modeling can further refine the process and enhance the precision of genome editing.
What are the potential limitations or drawbacks of the engineered pegRNA variants and prime-editor variants with M-MLV RT fused within SpCas9, and how can they be addressed?
While the fusion of M-MLV RT within SpCas9 in engineered pegRNA variants and prime-editor variants offers the advantage of spatial proximity and coordinated action during reverse transcription, there are potential limitations and drawbacks that need to be considered. One limitation could be the size and complexity of the fused protein, which may affect the delivery and expression efficiency in target cells. This could lead to reduced editing efficiency or off-target effects due to suboptimal protein folding or stability. Addressing this limitation may involve optimizing the fusion protein design to maintain the functionality of both M-MLV RT and SpCas9 while minimizing any negative impact on protein structure and function. Additionally, careful selection of the fusion site and linker sequences can help preserve the individual activities of M-MLV RT and SpCas9 within the fused protein. Continuous monitoring and characterization of the engineered variants through biochemical assays and cellular studies can provide insights into their performance and guide further optimization efforts.
What other structural or functional insights could be gained by studying the interactions between the prime editor components and the target DNA or cellular factors during the editing process?
Studying the interactions between the prime editor components and the target DNA or cellular factors during the editing process can yield valuable structural and functional insights that enhance our understanding of prime editing mechanisms. By investigating the dynamic conformational changes and molecular interactions within the SpCas9–M-MLV RTΔRNaseH–pegRNA–target DNA complex at different stages of editing, researchers can elucidate the precise coordination of protein activities and nucleic acid processing during prime editing. Furthermore, exploring the interplay between the prime editor components and cellular factors, such as chromatin structure or DNA repair machinery, can provide insights into the regulatory mechanisms that influence editing efficiency and specificity in different cellular contexts. Additionally, structural studies can reveal potential allosteric sites or interaction interfaces that can be targeted for modulating prime editing outcomes or developing novel editing strategies. By integrating structural and functional analyses, researchers can uncover key determinants of prime editing success and guide the rational design of improved prime editing tools for diverse genome engineering applications.
0
