Ubiquitin-Mediated Regulation of the Pattern-Recognition Receptor OsCERK1 Activates Immune Signaling in Rice
The article investigates the regulatory mechanisms that control the activation of the pattern-recognition receptor kinase OsCERK1, which is essential for plant immunity against microbial pathogens. The authors find that the U-box ubiquitin E3 ligase OsCIE1 acts as a molecular brake to inhibit OsCERK1 during homeostasis.
During normal conditions, OsCIE1 ubiquitinates OsCERK1, reducing its kinase activity and preventing autoimmunity. However, in the presence of the microbial pattern molecule chitin, active OsCERK1 phosphorylates OsCIE1, blocking its E3 ligase activity. This phosphorylation-dependent inactivation of OsCIE1 releases the brake on OsCERK1, allowing it to promote immune signaling and plant defense responses.
The authors further show that the phosphorylation of a specific serine residue within the U-box of OsCIE1 prevents its interaction with E2 ubiquitin-conjugating enzymes, serving as a phosphorylation switch that regulates the E3 ligase activity. Interestingly, this phosphorylation site is conserved in E3 ligases from plants to animals, suggesting a common regulatory mechanism.
The study identifies a ligand-released brake mechanism that enables dynamic and tightly controlled regulation of pattern-recognition receptor activation, which is crucial for effective immune responses while preventing autoimmunity.
Customize Summary
Rewrite with AI
Generate Citations
Translate Source
To Another Language
Generate MindMap
from source content
Visit Source
www.nature.com
Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice - Nature
Önemli Bilgiler Şuradan Elde Edildi
by Gang Wang,Xi... : www.nature.com 05-15-2024
https://www.nature.com/articles/s41586-024-07418-9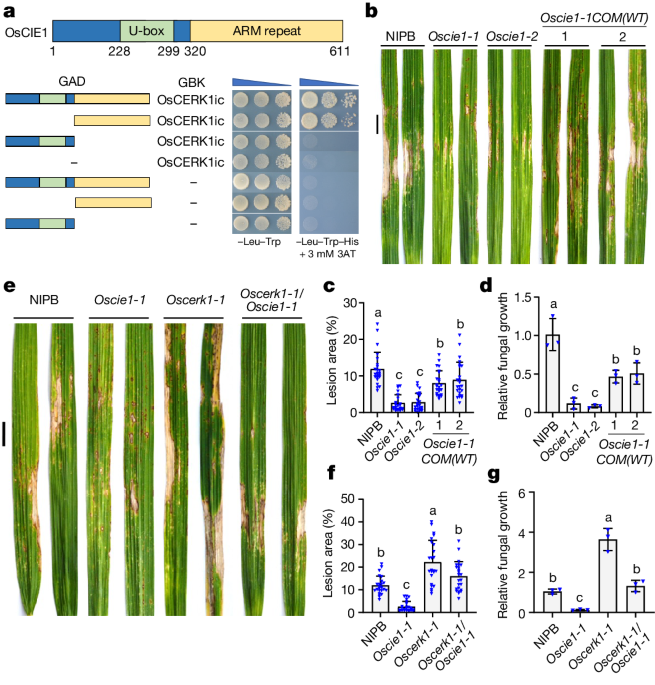
Daha Derin Sorular
