Structural Mechanism of mRNA Transcription Termination by Exoribonuclease
Core Concepts
Efficient mRNA transcription termination is facilitated by the exoribonuclease Rat1 in eukaryotic organisms.
Abstract
The content discusses the structural basis of mRNA transcription termination mediated by exoribonuclease in eukaryotic organisms, focusing on the mechanism involving Rat1 and its partner Rai1. The key highlights are:
- Eukaryotic organisms utilize exoribonuclease-mediated mechanism for mRNA transcription termination.
- Cryogenic electron microscopy structures of Saccharomyces cerevisiae Pol II pre-termination transcription complexes reveal the interaction between Rat1 and Pol II.
- Rat1 displaces the elongation factor Spt5 to dock at the Pol II stalk domain, guiding the nascent RNA towards its active center.
- Rat1 shields the RNA exit channel of Pol II and stacks three nucleotides at the 5′ terminus of the nascent RNA.
- Rat1 rotates towards Pol II as it shortens RNA, providing insights into the termination process.
- The findings offer a structural understanding of Rat1-mediated mRNA transcription termination in yeast and other eukaryotes.
Customize Summary
Rewrite with AI
Generate Citations
Translate Source
To Another Language
Generate MindMap
from source content
Visit Source
www.nature.com
Structural basis of exoribonuclease-mediated mRNA transcription termination - Nature
Stats
Eukaryotic organisms use a conserved exoribonuclease-mediated mechanism to terminate mRNA transcription by RNA polymerase II.
Rat1 displaces the elongation factor Spt5 to dock at the Pol II stalk domain.
Rat1 shields the RNA exit channel of Pol II, guides the nascent RNA towards its active center, and stacks three nucleotides at the 5′ terminus of the nascent RNA.
Rat1 rotates towards Pol II as it shortens RNA.
Quotes
"Our results provide the structural mechanism for the Rat1-mediated termination of mRNA transcription by Pol II in yeast and other eukaryotes."
Key Insights Distilled From
by Yuan Zeng,Ho... at www.nature.com 03-27-2024
https://www.nature.com/articles/s41586-024-07240-3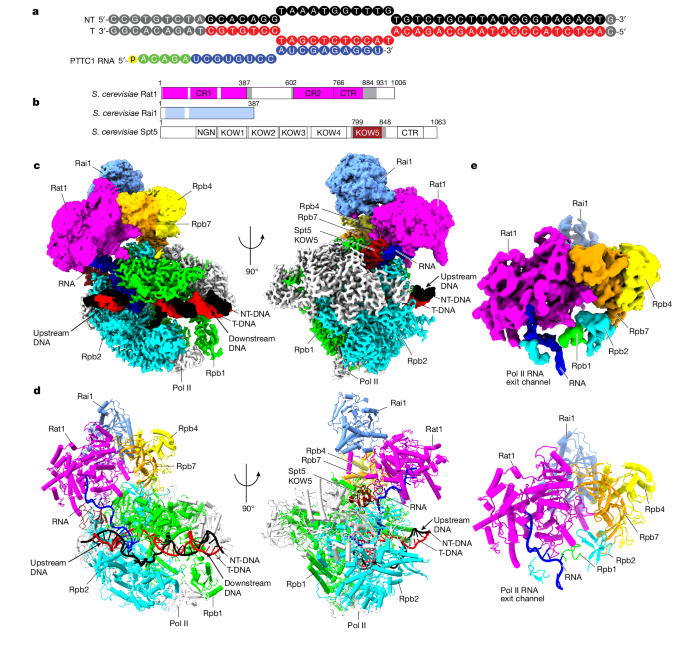
Deeper Inquiries
How does the structural mechanism of mRNA transcription termination by Rat1 in eukaryotic organisms compare to prokaryotic organisms
In eukaryotic organisms, the structural mechanism of mRNA transcription termination by Rat1 involves the displacement of the elongation factor Spt5 to dock at the Pol II stalk domain. Rat1 shields the RNA exit channel of Pol II, guides the nascent RNA towards its active center, and stacks three nucleotides at the 5′ terminus of the nascent RNA. As Rat1 shortens RNA, it rotates towards Pol II. This mechanism ensures efficient termination of mRNA transcription. In contrast, prokaryotic organisms utilize different termination mechanisms, such as the Rho-dependent and Rho-independent termination. Rho-dependent termination involves the Rho protein binding to the nascent RNA and causing transcription termination, while Rho-independent termination relies on specific sequences in the RNA forming secondary structures that lead to termination. The structural differences in termination mechanisms between eukaryotic Rat1-mediated termination and prokaryotic termination mechanisms highlight the evolutionary divergence in transcription termination strategies between these two domains of life.
What potential implications could understanding exoribonuclease-mediated termination have on gene expression regulation
Understanding exoribonuclease-mediated termination, such as the mechanism involving Rat1 in eukaryotic organisms, can have significant implications on gene expression regulation. By elucidating the structural basis of how Rat1 terminates mRNA transcription by Pol II, researchers can gain insights into the precise molecular events that govern this crucial process. This knowledge can lead to the development of targeted therapies that modulate gene expression by manipulating transcription termination. Additionally, understanding how exoribonucleases like Rat1 function in gene regulation can provide valuable information for studying diseases linked to transcription dysregulation, such as cancer. Insights into the termination of mRNA transcription can also shed light on how cells control gene expression levels and respond to environmental cues, offering new avenues for research in molecular biology and biotechnology.
How might advancements in cryogenic electron microscopy further enhance our understanding of transcription termination mechanisms
Advancements in cryogenic electron microscopy (cryo-EM) can further enhance our understanding of transcription termination mechanisms by providing high-resolution structural insights into the molecular interactions involved. Cryo-EM allows researchers to visualize macromolecular complexes, such as the Pol II pre-termination transcription complexes bound to Rat1 and Rai1, in their native state. By capturing these complexes at near-atomic resolution, cryo-EM can reveal the precise spatial arrangement of proteins, RNA, and DNA during transcription termination. This detailed structural information can help elucidate the dynamic conformational changes that occur during termination and provide a molecular basis for understanding the functional roles of key players like Rat1. Moreover, cryo-EM can facilitate the visualization of transient intermediates and conformational states that are challenging to capture using other structural biology techniques, offering a comprehensive view of the termination process at the molecular level.
0
