insight - Computational Biology - # Tracing the Historical Spread of Malaria Parasites Through Ancient Genomic Analysis
Ancient Plasmodium Genomes Reveal the Spread and History of Human Malaria Across the Globe
Core Concepts
Ancient genomic analysis of Plasmodium parasites reveals the long-standing presence and global dissemination of human malaria, driven by cross-cultural contacts and population movements.
Abstract
The article presents findings from a comprehensive ancient genomic analysis of Plasmodium parasites, the causative agents of human malaria. The researchers generated high-coverage mitochondrial and nuclear genome data from P. falciparum, P. vivax, and P. malariae samples spanning around 5,500 years of human history across 16 countries.
Key insights:
The evidence indicates that P. vivax and P. falciparum were present in geographically disparate regions of Eurasia as early as the 4th and 1st millennia BCE, respectively, predating textual references by several millennia.
Genomic analysis suggests distinct disease histories for P. falciparum and P. vivax in the Americas: European colonizers were the source of American P. vivax, while the trans-Atlantic slave trade likely introduced P. falciparum.
The data underscores the role of cross-cultural contacts and population movements in the dissemination of malaria, providing a biomolecular foundation for future research on the historical impact of Plasmodium parasites on human populations.
The unexpected discovery of P. falciparum in the high-altitude Himalayas offers a rare case study where individual mobility can be inferred from infection status, shedding light on cross-cultural connectivity in the region nearly three millennia ago.
Ancient Plasmodium genomes shed light on the history of human malaria - Nature
Stats
The study analyzed Plasmodium genomes from samples spanning around 5,500 years of human history across 16 countries.
The evidence indicates the presence of P. vivax and P. falciparum in Eurasia as early as the 4th and 1st millennia BCE, respectively.
Quotes
"Genomic analysis supports distinct disease histories for P. falciparum and P. vivax in the Americas: similarities between now-eliminated European and peri-contact South American strains indicate that European colonizers were the source of American P. vivax, whereas the trans-Atlantic slave trade probably introduced P. falciparum into the Americas."
"Our unexpected discovery of P. falciparum in the high-altitude Himalayas provides a rare case study in which individual mobility can be inferred from infection status, adding to our knowledge of cross-cultural connectivity in the region nearly three millennia ago."
Key Insights Distilled From
by Megan Michel... at www.nature.com 06-12-2024
https://www.nature.com/articles/s41586-024-07546-2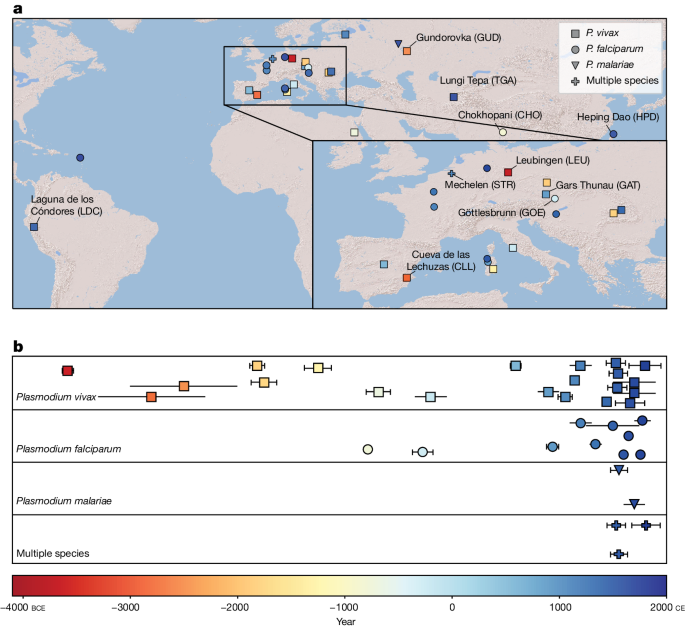
Deeper Inquiries
How might the historical spread of malaria parasites have influenced the genetic diversity and evolution of human populations?
The historical spread of malaria parasites could have significantly influenced the genetic diversity and evolution of human populations through the process of natural selection. As mentioned in the context, malaria-causing protozoa like Plasmodium have exerted strong selective pressures on the human genome. This selective pressure has led to the emergence of resistance alleles in human populations living in malaria-endemic regions. Over time, individuals with these resistance alleles would have had a survival advantage, leading to the spread of these beneficial genetic variations within the population. This process, known as positive selection, can result in changes in the frequency of certain genetic variants in a population, ultimately contributing to genetic diversity and evolution.
What other ancient pathogens could be studied using similar ancient genomic approaches to uncover their historical impact on human societies?
Similar ancient genomic approaches could be applied to study other ancient pathogens such as tuberculosis, leprosy, smallpox, and influenza. By analyzing ancient DNA from skeletal remains or archaeological samples, researchers can uncover the genetic makeup of these pathogens and trace their historical spread and impact on human societies. For example, studying the ancient genomes of tuberculosis strains could provide insights into the evolution of the disease and its transmission patterns in different populations over time. Similarly, analyzing the genomes of ancient smallpox viruses could shed light on the history of the disease and its role in shaping human populations.
What insights could be gained by comparing the ancient genomic data of Plasmodium parasites with historical records and archaeological evidence of human migration and cross-cultural interactions?
Comparing the ancient genomic data of Plasmodium parasites with historical records and archaeological evidence of human migration and cross-cultural interactions can provide valuable insights into the spread of malaria and its impact on human populations throughout history. By correlating genetic data with historical information, researchers can reconstruct the movement of ancient human populations and the transmission of malaria parasites across different regions. This comparative analysis can help identify key events, such as trade routes, colonization efforts, or population movements, that facilitated the spread of malaria. Additionally, studying the genetic diversity of Plasmodium parasites in conjunction with archaeological evidence can offer a more comprehensive understanding of how human activities and interactions have influenced the epidemiology of malaria over time.
0
