insight - Computational Biology - # Chromatin Accessibility and Transcriptional Regulation in Early Human Neurodevelopment
Comprehensive Analysis of Chromatin Accessibility and Gene Expression Dynamics During Early Human Brain Development in the First Trimester
Core Concepts
Chromatin accessibility and gene expression patterns across the entire developing human brain during the first trimester (6–13 weeks after conception) provide insights into key gene regulatory mechanisms underlying the emergence of brain cell types.
Abstract
The content describes a comprehensive analysis of chromatin accessibility and paired gene expression across the entire developing human brain during the first trimester (6–13 weeks after conception). The key highlights and insights are:
The researchers defined 135 clusters and used multi-omic measurements to link candidate cis-regulatory elements to gene expression.
The number of accessible regions increased with age and along neuronal differentiation.
Using a convolutional neural network, the researchers identified putative functional transcription factor-binding sites in enhancers characterizing neuronal subtypes.
They applied this model to cis-regulatory elements linked to ESRRB to elucidate its activation mechanism in the Purkinje cell lineage.
By linking disease-associated single nucleotide polymorphisms to cis-regulatory elements, the researchers validated putative pathogenic mechanisms in several diseases and identified midbrain-derived GABAergic neurons as being the most vulnerable to major depressive disorder-related mutations.
The findings provide a comprehensive reference for future studies related to human neurodevelopment and offer insights into key gene regulatory mechanisms underlying the emergence of brain cell types during the first trimester.
Chromatin accessibility during human first-trimester neurodevelopment - Nature
Stats
We defined 135 clusters and used multiomic measurements to link candidate cis-regulatory elements to gene expression.
The number of accessible regions increased both with age and along neuronal differentiation.
Quotes
"Using a convolutional neural network, we identified putative functional transcription factor-binding sites in enhancers characterizing neuronal subtypes."
"By linking disease-associated single nucleotide polymorphisms to cis-regulatory elements, we validated putative pathogenic mechanisms in several diseases and identified midbrain-derived GABAergic neurons as being the most vulnerable to major depressive disorder-related mutations."
Key Insights Distilled From
by Camiel C. A.... at www.nature.com 05-01-2024
https://www.nature.com/articles/s41586-024-07234-1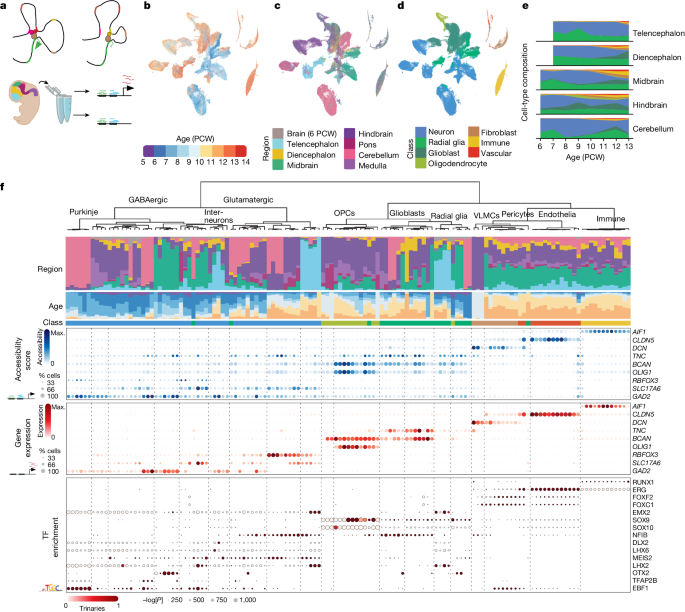
Deeper Inquiries
How can the insights from this study be leveraged to develop novel therapeutic interventions for neurodevelopmental disorders?
The insights from this study on chromatin accessibility and gene expression during early human brain development can be instrumental in developing novel therapeutic interventions for neurodevelopmental disorders. By understanding the regulatory mechanisms underlying the emergence of different brain cell types, researchers can target specific transcription factors or cis-regulatory elements associated with neurodevelopmental disorders. For example, identifying enhancers characterizing neuronal subtypes through the convolutional neural network can lead to the development of targeted therapies that modulate these specific regulatory elements to correct abnormalities in neuronal development. Additionally, linking disease-associated single nucleotide polymorphisms to cis-regulatory elements can provide insights into the pathogenic mechanisms of neurodevelopmental disorders, enabling the development of precision medicine approaches tailored to individual genetic profiles.
What are the potential limitations of the convolutional neural network approach used to identify transcription factor-binding sites, and how could it be further improved?
While the convolutional neural network approach is powerful in identifying putative functional transcription factor-binding sites in enhancers, it has certain limitations that need to be considered. One limitation is the reliance on the quality and quantity of training data, as the accuracy of the model is highly dependent on the diversity and representativeness of the input data. Additionally, the interpretability of the model outputs may be challenging, making it difficult to understand the underlying biological mechanisms driving the predictions. To address these limitations, the convolutional neural network approach could be further improved by incorporating more diverse and comprehensive datasets of transcription factor-binding sites to enhance the model's predictive accuracy. Moreover, integrating additional layers of interpretability, such as attention mechanisms or feature visualization techniques, can help researchers understand how the model makes predictions and provide insights into the biological relevance of the identified transcription factor-binding sites.
What other types of multi-omic data could be integrated with chromatin accessibility and gene expression to provide a more comprehensive understanding of early human brain development?
To gain a more comprehensive understanding of early human brain development, researchers can integrate additional types of multi-omic data with chromatin accessibility and gene expression data. One valuable dataset to incorporate is DNA methylation data, which can provide insights into epigenetic modifications that regulate gene expression patterns during neurodevelopment. By analyzing DNA methylation profiles in conjunction with chromatin accessibility and gene expression data, researchers can uncover the epigenetic landscape of the developing brain and identify key regulatory elements involved in neurodevelopmental processes. Furthermore, integrating single-cell RNA sequencing data can offer a detailed characterization of cell types and states within the developing brain, allowing for the identification of cell-specific regulatory networks and interactions. By combining chromatin accessibility, gene expression, DNA methylation, and single-cell RNA sequencing data, researchers can obtain a holistic view of the molecular mechanisms governing early human brain development.
0
