Targeted In Situ Base Editing of Gut Bacteria in Mice Using Engineered Phage Particles
Core Concepts
Engineered phage-derived particles can efficiently deliver base editors to modify target genes in gut bacteria colonizing the mouse intestine, enabling investigation of bacterial gene function and potential microbiome-targeted therapies.
Abstract
The article describes the development of a novel approach to edit bacterial genomes directly in the mouse gut using engineered phage-derived particles. The key highlights are:
- Researchers engineered phage-derived particles to deliver a base editor and modify Escherichia coli colonizing the mouse gut.
- Editing of a β-lactamase gene in a model E. coli strain resulted in a median editing efficiency of 93% of the target bacterial population with a single dose.
- The edited bacteria were stably maintained in the mouse gut for at least 42 days following treatment, achieved using a non-replicative DNA vector to prevent maintenance and dissemination of the payload.
- The researchers then leveraged this approach to edit several genes of therapeutic relevance in E. coli and Klebsiella pneumoniae strains in vitro and demonstrate in situ editing of a gene involved in the production of curli in a pathogenic E. coli strain.
- This work demonstrates the feasibility of directly modifying bacteria in the gut, offering a new avenue to investigate the function of bacterial genes and opening the door to the design of new microbiome-targeted therapies.
Customize Summary
Rewrite with AI
Generate Citations
Translate Source
To Another Language
Generate MindMap
from source content
Visit Source
www.nature.com
In situ targeted base editing of bacteria in the mouse gut - Nature
Stats
Median editing efficiency of 93% of the target bacterial population with a single dose.
Edited bacteria were stably maintained in the mouse gut for at least 42 days following treatment.
Quotes
"Editing of a β-lactamase gene in a model E. coli strain resulted in a median editing efficiency of 93% of the target bacterial population with a single dose."
"Edited bacteria were stably maintained in the mouse gut for at least 42 days following treatment."
Key Insights Distilled From
by Andr... at www.nature.com 07-10-2024
https://www.nature.com/articles/s41586-024-07681-w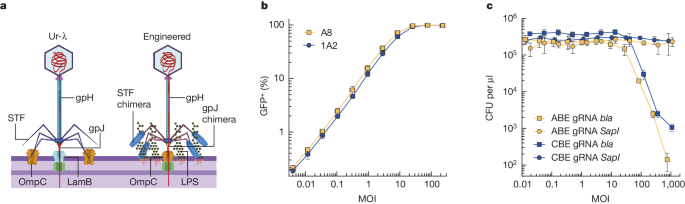
Deeper Inquiries
How could this in situ bacterial genome editing approach be leveraged to modulate the gut microbiome for therapeutic purposes, such as treating dysbiosis or targeting pathogenic bacteria?
This in situ bacterial genome editing approach offers a promising avenue for modulating the gut microbiome to address various therapeutic goals. By precisely editing the genomes of specific bacterial strains in the gut, it becomes possible to target pathogenic bacteria responsible for infections or dysbiosis. For instance, by editing genes involved in virulence factors or antibiotic resistance mechanisms, the pathogenicity of harmful bacteria can be reduced, leading to improved health outcomes. Additionally, this approach could be utilized to enhance the presence of beneficial bacteria in the gut by editing genes that promote beneficial traits such as the production of beneficial metabolites or the inhibition of pathogen colonization. Overall, the ability to selectively modify bacterial populations in the gut opens up new possibilities for developing targeted therapies to restore microbial balance and combat various gut-related disorders.
What are the potential limitations or challenges of this approach, such as off-target effects, immune responses, or the ability to edit a broader range of bacterial species?
While in situ bacterial genome editing holds great promise, there are several potential limitations and challenges that need to be addressed. One significant concern is the possibility of off-target effects, where unintended edits occur in the bacterial genome, leading to unpredictable outcomes. To mitigate this risk, rigorous testing and optimization of the editing system are essential to ensure high specificity and accuracy. Another challenge is the potential for immune responses triggered by the delivery of editing tools, which could impact the effectiveness of the approach. Strategies to minimize immune reactions, such as using biocompatible delivery vehicles or optimizing dosing regimens, will be crucial for the success of this technology. Furthermore, the current approach may have limitations in editing a broader range of bacterial species beyond Escherichia coli and Klebsiella pneumoniae. Developing versatile editing tools that can target a wider spectrum of bacterial species will be essential for the widespread application of this technology in modulating the gut microbiome for therapeutic purposes.
What other applications beyond investigating bacterial gene function could this technology enable, such as engineering beneficial bacteria or studying host-microbiome interactions?
Beyond investigating bacterial gene function, this technology opens up a range of exciting applications with significant implications for human health and biotechnology. One key application is the engineering of beneficial bacteria with enhanced functionalities for therapeutic purposes. By editing genes involved in producing beneficial metabolites, enhancing probiotic properties, or modulating immune responses, it is possible to design custom bacterial strains tailored for specific health benefits. Moreover, this technology can facilitate the study of host-microbiome interactions by enabling precise manipulation of bacterial genes involved in crosstalk with the host. By editing genes that influence host immune responses, metabolism, or signaling pathways, researchers can gain insights into the complex interplay between the microbiome and host physiology. Overall, the ability to engineer bacterial genomes in situ offers a powerful tool for advancing our understanding of microbial communities and developing innovative solutions for human health and biotechnology.
0
