insight - Computational Biology - # Spatial Organization of Neuronal Transcriptomic Profiles in the Cerebral Cortex
Whole-Cortex In Situ Sequencing Reveals How Neuronal Transcriptomic Signatures Reflect Cortical Area Identity and Connectivity
Core Concepts
Cortical areas with distinct cytoarchitecture, connectivity, and neuronal activity are reflected in the transcriptomic signatures of individual neurons, which are shaped by peripheral sensory inputs during development.
Abstract
The study used a high-throughput in situ sequencing technique called BARseq to interrogate the expression of 104 cell-type marker genes in over 10 million cells, including 4.2 million cortical neurons, across nine mouse forebrain hemispheres. The key findings are:
De novo clustering of gene expression in single neurons revealed transcriptomic types consistent with previous single-cell RNA sequencing studies.
The composition of these transcriptomic types is highly predictive of cortical area identity, suggesting that the spatial organization of the cortex is reflected in neuronal transcriptomic signatures.
Cortical areas with similar compositions of transcriptomic types, defined as cortical modules, overlap with areas that are highly connected, indicating that the same modular organization is reflected in both transcriptomic signatures and connectivity.
Neonatal binocular enucleation (removal of both eyes) caused the shifting of cell-type compositional profiles of visual areas towards neighboring cortical areas within the same module, suggesting that peripheral sensory inputs help sharpen the distinct transcriptomic identities of areas within cortical modules during development.
The study provides a proof of principle for using large-scale in situ sequencing to reveal the brain-wide molecular architecture and understand its development.
Whole-cortex in situ sequencing reveals input-dependent area identity - Nature
Stats
The study analyzed the expression of 104 cell-type marker genes in 10.3 million cells, including 4,194,658 cortical neurons, across nine mouse forebrain hemispheres.
Neonatal binocular enucleation was used to assess the impact of peripheral sensory inputs on the development of cortical transcriptomic signatures.
Quotes
"The composition of transcriptomic types is highly predictive of cortical area identity."
"Cortical areas with similar compositions of transcriptomic types, which we defined as cortical modules, overlap with areas that are highly connected, suggesting that the same modular organization is reflected in both transcriptomic signatures and connectivity."
"Binocular enucleation caused the shifting of the cell-type compositional profiles of visual areas towards neighbouring cortical areas within the same module, suggesting that peripheral inputs sharpen the distinct transcriptomic identities of areas within cortical modules."
Key Insights Distilled From
by Xiaoyin Chen... at www.nature.com 04-24-2024
https://www.nature.com/articles/s41586-024-07221-6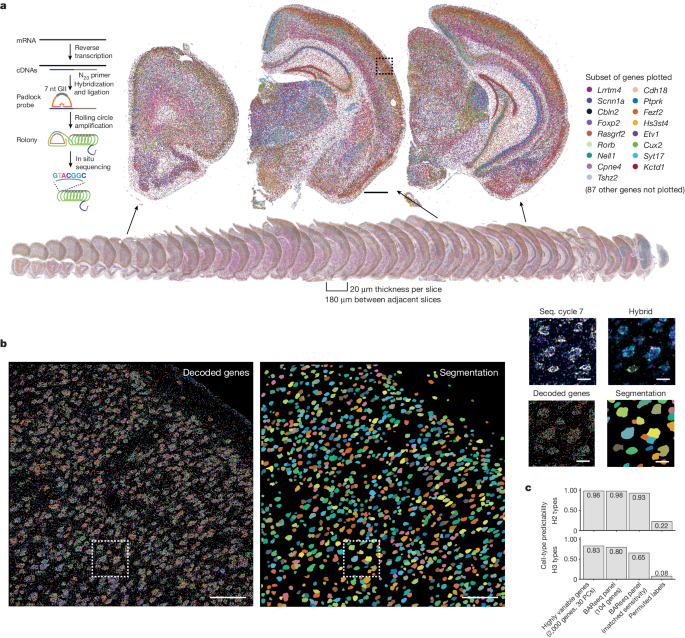
Deeper Inquiries
How do the transcriptomic signatures of cortical neurons relate to their functional properties and information processing capabilities?
The transcriptomic signatures of cortical neurons play a crucial role in determining their functional properties and information processing capabilities. The diverse gene expression patterns observed in different neuronal types contribute to the specialization of cortical areas, each with unique cytoarchitecture, connectivity, and activity patterns. These transcriptomic signatures are reflective of the specific functions that neurons within a particular cortical area perform. For example, neurons in visual areas may express genes related to visual processing, while neurons in motor areas may have gene expression patterns associated with motor control. By analyzing the transcriptomic profiles of cortical neurons, researchers can gain insights into the molecular basis of neuronal function and how information is processed within different cortical regions.
What other developmental or environmental factors, besides sensory inputs, might shape the establishment of distinct transcriptomic identities of cortical areas?
In addition to sensory inputs, several other developmental and environmental factors can influence the establishment of distinct transcriptomic identities of cortical areas. One such factor is genetic regulation, where the expression of certain genes during development can shape the transcriptomic profiles of cortical neurons. Epigenetic mechanisms, such as DNA methylation and histone modifications, can also play a role in determining gene expression patterns in different cortical areas. Environmental factors, such as exposure to stress or enriched environments, can impact gene expression and contribute to the diversity of transcriptomic signatures observed in the cortex. Additionally, interactions with other brain regions and neural circuits during development can influence the transcriptomic identities of cortical areas, highlighting the complex interplay between genetic, environmental, and neural factors in shaping the molecular architecture of the brain.
Could the insights from this study on the spatial organization of the cerebral cortex be extended to understand the molecular architecture and development of other complex brain regions?
The insights gained from studying the spatial organization of the cerebral cortex using in situ sequencing techniques can indeed be extended to understand the molecular architecture and development of other complex brain regions. The modular organization of the cortex, where areas with similar transcriptomic signatures are highly connected, suggests a common organizational principle that may be present in other brain regions. By applying similar high-throughput sequencing approaches to different brain regions, researchers can uncover the transcriptomic diversity and connectivity patterns that underlie the functional specialization of these regions. Understanding how gene expression profiles contribute to the development and organization of complex brain regions beyond the cortex can provide valuable insights into the molecular mechanisms that govern brain function and behavior.
0
